Simple Tutorial¶
I recommend running this using a jupyter notebook.
Alternatively use a google colab.
notebook to get started quickly. (Use !pip install kinetics in the first cell)
Or here is a direct link to this workbook in google colab. https://colab.research.google.com/github/willfinnigan/kinetics/blob/master/examples/Simple_example.ipynb
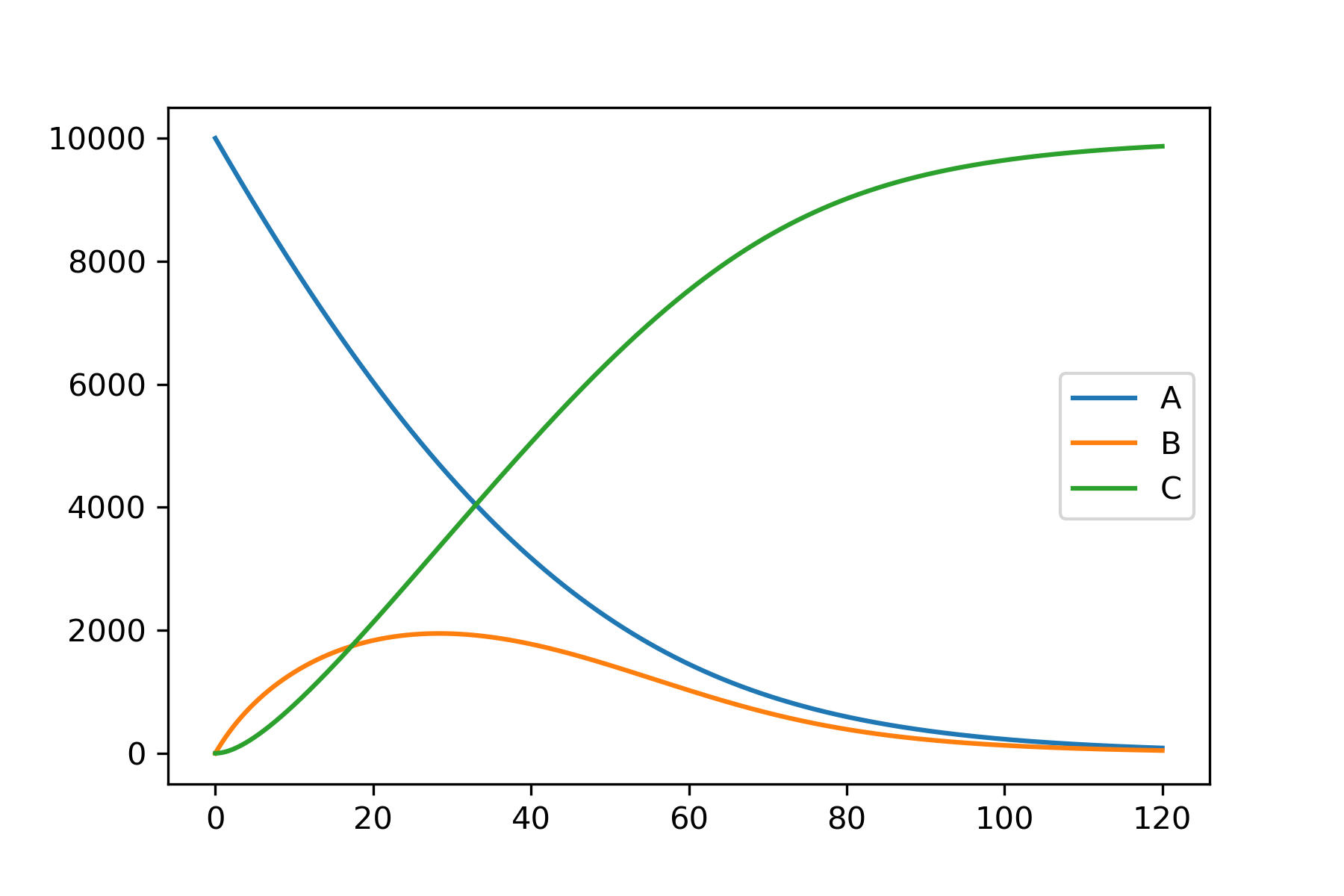
This example shows the code required to model the relatively simple system shown on the main page. The complete example as a single block of code is available at the end.
Complete code example¶
Below is the complete code for this example. Copy it into a jupyter notebook or google colab notebook to get started. This example is then explained step by step in the rest of this tutorial.
# Uncomment and run this if using google colab
# !pip install kinetics
import kinetics
import matplotlib.pyplot as plt
%config InlineBackend.figure_format ='retina'
# Define reactions
enzyme_1 = kinetics.Uni(kcat='enz1_kcat', kma='enz1_km', enz='enz_1', a='A',
substrates=['A'], products=['B'])
enzyme_1.parameters = {'enz1_kcat' : 100,
'enz1_km' : 8000}
enzyme_2 = kinetics.Uni(kcat='enz2_kcat', kma='enz2_km', enz='enz_2', a='B',
substrates=['B'], products=['C'])
enzyme_2.parameters = {'enz2_kcat' : 30,
'enz2_km' : 2000}
# Set up the model
model = kinetics.Model(logging=False)
model.append(enzyme_1)
model.append(enzyme_2)
model.set_time(0, 120, 1000) # 120 mins, 1000 timepoints.
# Set starting concentrations
model.species = {"A" : 10000,
"enz_1" : 4,
"enz_2" : 10}
model.setup_model()
# Run the model
model.run_model()
model.plot_substrate('A')
model.plot_substrate('B')
model.plot_substrate('C', plot=True)
# Now try altering the enzyme concentration, km or kcat, and re-running the model to see the effects this has....
Define reactions¶
The first step in building a kinetic model using this package is to define rate equations which describe the rates of the reactions in a system. Rate equations will typically contain rate constants, referred to as parameters here, and species concentrations. Each rate equation which needs to be included in the model should be set up as a reaction object, as shown below.
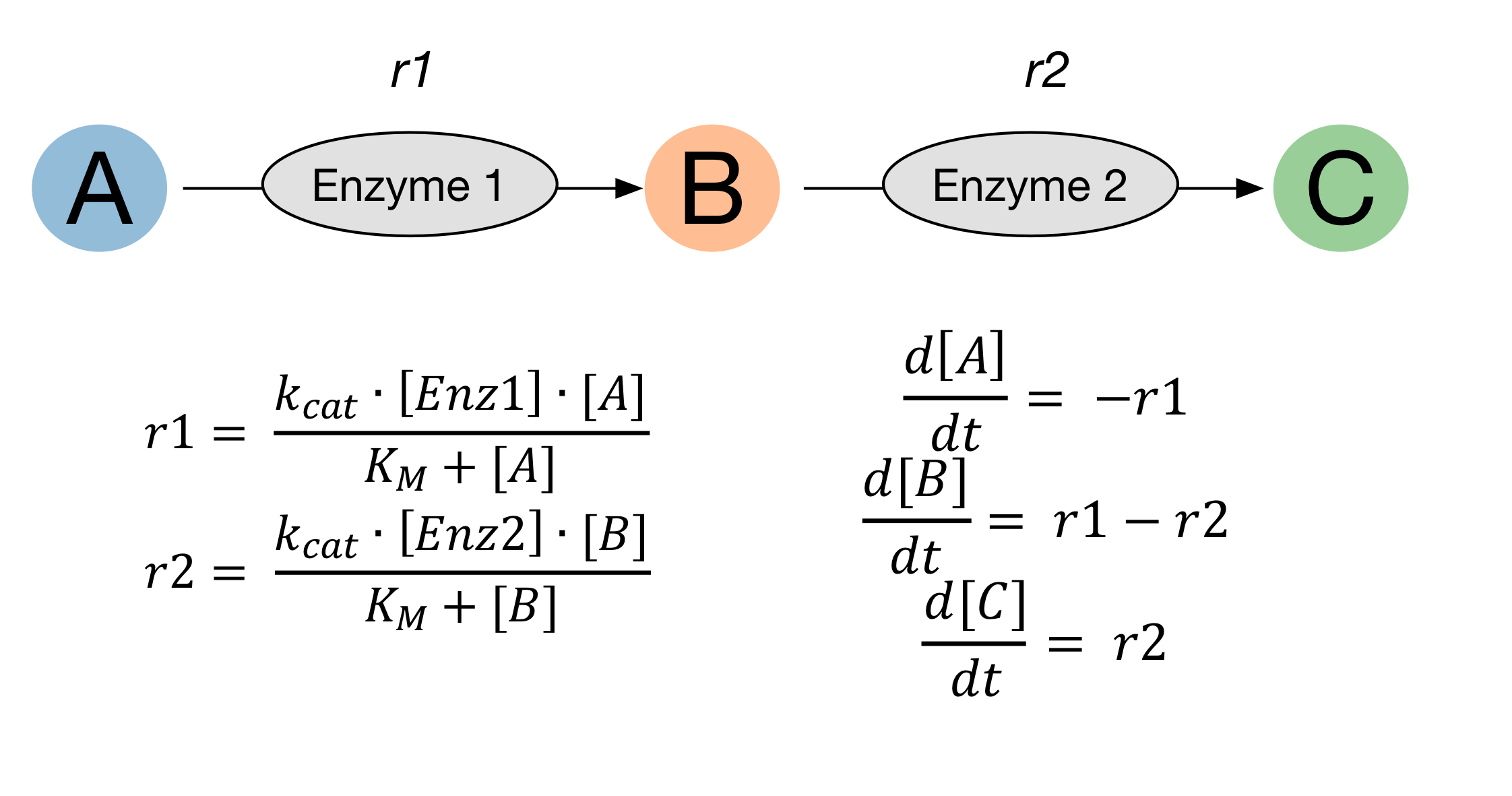
In this example, a pair of single substrate irreversible enzyme reactions which follow Michaelis-Menton kinetics are shown:
import kinetics
enzyme_1 = kinetics.Uni(kcat='enz1_kcat', kma='enz1_km',
enz='enz_1', a='A',
substrates=['A'], products=['B'])
enzyme_1.parameters = {'enz1_kcat' : 100,
'enz1_km' : 8000}
enzyme_2 = kinetics.Uni(kcat='enz2_kcat', kma='enz2_km',
enz='enz_2', a='B',
substrates=['B'], products=['C'])
enzyme_2.parameters = {'enz2_kcat' : 30,
'enz2_km' : 2000}
When setting up reactions, parameter names (kcat='enz1_kcat', kma='enz1_km') and species names (enz='enz_1', a='A',) are first given.
These are used by the reaction when calculating the rate.
A list of substrates which are used up in the reaction, and products that are made in the reaction is the specified (substrates=['A'], products=['B']).
Most of the common enzyme mechanisms are pre-defined in the package. For more information see Reactions (link).
Next we need to add these reactions to a model.
Define the model¶
The model class is central to the kinetics package. It is essentially a list to which we append our reactions, with some extra variables and functions to run the model.
import kinetics
# Initiate a new model
model = kinetics.Model()
Add reactions to the model¶
The reactions we defined above are appended to the model.
# Append our reactions.
model.append(enzyme_1)
model.append(enzyme_2)
Set how long the model will simulate¶
The time that a model will simulate can be set by using:
# Set the model to run from 0 to 120 minutes, over 1000 steps
model.set_time(0, 120, 1000)
Set starting species concentrations¶
The model defaults all starting species to 0, for anything defined in the reactions. For the model to predict anything useful, we need to give it starting concentrations (including the enzymes). Only species which are greater than 0 need to be defined here.
# Set starting concentrations
model.species = {"A" : 10000,
"enz_1" : 4,
"enz_2" : 10}
Run the model¶
Once everything is set, run model_one.setup_model() followed by model_one.run_model().
A dataframe containing the simulation results is then available using model_one.results_dataframe().
Alternatively, results can plotted directly using an in-built plot function model_one.plot_substrate('A').
# Setup and run the model
model.setup_model()
model.run_model()
# Plot the results
model.plot_substrate('A')
model.plot_substrate('B')
model.plot_substrate('C', plot=True)